Bash Buffoonery - Slimming down piped commands with awk
Creating some of the most convoluted bash commands for fun!
 {: .yes}
{: .yes}
Let’s set the scene:
I am working on a .tf file that would be responsible for provisioning some infrastructure with OpenTofu. But instead of just directly filling out all the values (and then forgetting about it and accidentally committing them on a repo), I want to try to do some Terraform Best Practices. One best practice was to segment the files, this is done by splitting off the main file into varous other files and then putting the sensitive values on a .tfvar file.
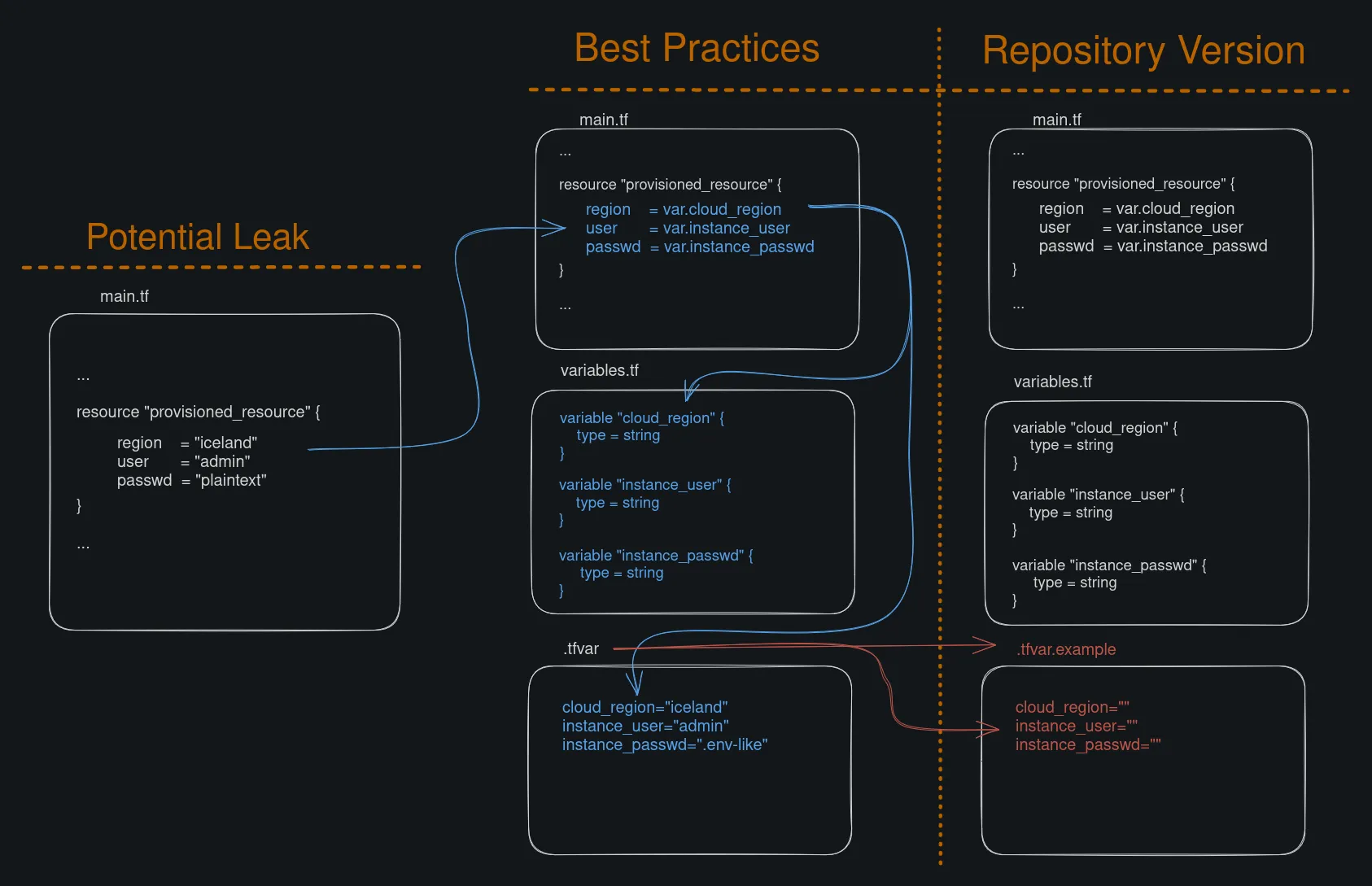
Effectively, by splitting the main.tf file into main.tf, variables.tf, and .tfvar in additional to putting .tfvar as a line in the .gitignore file, accidentally leaking the credentials should be no concern.
Another helpful thing is to have is an example of what a valid .tfvar file might look like (perhaps call it .tfvar.example). Which sounds like something that can get neglected very easily, if there wasn’t a trivial way of automating it and a documented process. When it comes to automating stuff, this sounds like a perfect time for some bash buffoonery! (A blatant disregard for documentation could be considered just plain buffoonery.)
So the goal is to create a valid .tfvar.example file from an existing variables.tf file and make it as compact as possible to write (and maybe more performant, maybe).
Take the following file as what a variables.tf file might look like:
# Maybe a comment
variable "tomatoes" {
type = string
}
# Maybe some documentation
variable "potatoes" {
type = string
}
# toma toma porfa
variable "chicha-morada" {
type = string
}
...
With this in mind, let’s see that we can do!
$ grep -E 'variable' variables.tf | awk '{print $2}' | tr -d '"'
tomatoes
potatoes
chicha-morada
Cool! The variables were displayed, however it might be more useful to have it in the format where it isn’t tomatoes but rather tomatoes=“”(Back to the drawing board)
Code Breakdown:
grep -E 'variable' variables.tf→ search all lines that contain the word variableawk '{print $2}'→ display 2nd column to screen, by defaultawkuses SPACE as a delimitertr -d '"'→ removes all instances of"
$ for l in `grep -E 'variable' variables.tf | awk '{print $2}' | tr -d '"'`; do echo "$l=\"\""; done
tomatoes=""
potatoes=""
chicha-morada=""
Okay, not bad but I don’t want to see that I am doing a loop. Can some length also be pared?
Code Breakdown:
for l in <CMD>; do→ iterate line by line via the previous command asecho "$l=\"\""→ for each line being iterated, display the line with a=""at the end
$ grep -E 'variable' variables.tf | awk '{print $2}' | tr -d '"' | sed -E 's/(.*)/\1=""/g'
tomatoes=""
potatoes=""
chicha-morada=""
Okay, less text so nice but there are a couple problems here. One grep is returning any line that contains the word variable, only lines starting with the word variable would be best. Additionally, invoking the (.*) within the sed section is contentious, while it isn’t a textbook example of evil regex, improvements can be made to make it less ambiguous and more strict.
If curious about a real life example of the regex denial of service (ReDoS) check out Kevin Fang’s video about Cloudflare. Cloudflare also has a brilliant blog on the outage as well as blog.
Code Breakdown:
sed -E 's/(.*)/\1=""/g→ Does a lot with little typing!
-E→ uses extended regular expressions in the script (necessary for back-references)s/a/b/g→ do a globalgstring substitutionswhich turnsaintob(and)→ is a capture group, and with looks for any content.0 or more times*\1→ back-reference to our first capture group and then add a=""to the end of all the things found.
$ cat variables.tf | awk '/^variable/{gsub("\"",""); print $2 "=\"\""}'
tomatoes=""
potatoes=""
chicha-morada=""
awk has now become the show-stealer of this bash script! It may be faster to run awk directly on the file as opposed to piped in. (But also curious if that is a valid claim)
Code Breakdown:
cat variables.tf→ show thevariables.tffileawk→ It does a lot:
/^variable/→ filters for lines that start with the wordvariablegsub("\"","");→ removes all the instances of"print $2 "=\"\""→ responsible for adding the=""to the end of the line
$ awk '/^variable/{gsub("\"",""); print $2 "=\"\""}' variables.tf
tomatoes=""
potatoes=""
chicha-morada=""
Epic! Okay, but can content also be filtered out? (Asking for curiosity sake)
Code Breakdown:
- Removed the
cat variables.tfpipe to just directly read thevariable.tffile
$ awk '/^variable/ && !/morada/ {gsub("\"",""); print $2 "=\"\""}' variables.tf
tomatoes=""
potatoes=""
$ awk '/^variable/ && !/morada/ && $2 {gsub("\"",""); print $2 "=\"\""}' variables.tf
# ensures that no empty lines get picked up
tomatoes=""
potatoes=""
Dope! Filtering both inclusively and exclusively has been achieved!
(A better use might be to exclude lines that start with a comment or a line saying TODO)
Also, the second command checks that the second field exists (so writing a bunch lines starting with the word variable and then forgetting about their existence isn’t gonna create any ‘spooky’ phantom lines).
Code Breakdown:
&& !/morada/adds another conditional which excludes any line containing the wordmorada$2→ checks that the 2nd column exists and is not null
Conclusion
# try-1.sh --> 100 (character count)
for l in $(grep -E 'variable' variables.tf | awk '{print $2}' | tr -d '"'); do echo "$l=\"\""; done
# try-2.sh --> 89 (character count)
grep -E 'variable' variables.tf | awk '{print $2}' | tr -d '"' | sed -E 's/(.*)/\1=""/g'
# try-3.sh --> 70 (character count)
cat variables.tf | awk '/^variable/{gsub("\"",""); print $2 "=\"\""}'
# try-4.sh --> 64 (character count)
awk '/^variable/{gsub("\"",""); print $2 "=\"\""}' variables.tf
Automation with awk has been a success. As a result of relying on awk, the initial bash command that took 100 characters to write, now takes only 64, while retaining functionality. Also, now some deeper knowledge of awk has been attained. However, where does that leave performance? Which command might be best suitable for speed?
Performance - A Quick and Dirty Test
(*Read with caution)

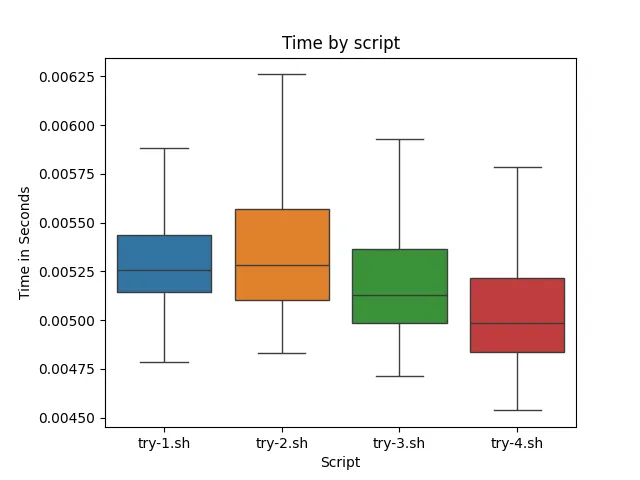
For testing the commands, I am using perf stat for more precision than time. Additionally, I am running the command 10000 times and just doing a basic 5-number summary on the value of the line displaying seconds time elapsed, so (I can stress this enough) take this data with a grain of salt.
# try-1.sh
for l in $(grep -E 'variable' variables.tf | awk '{print $2}' | tr -d '"'); do echo "$l=\"\""; done
# try-2.sh
grep -E 'variable' variables.tf | awk '{print $2}' | tr -d '"' | sed -E 's/(.*)/\1=""/g'
# try-3.sh
cat variables.tf | awk '/^variable/{gsub("\"",""); print $2 "=\"\""}'
# try-4.sh
awk '/^variable/{gsub("\"",""); print $2 "=\"\""}' variables.tf
# Stats
try-1.sh try-2.sh try-3.sh try-4.sh
count 10000.000000 10000.000000 10000.000000 10000.000000
mean 0.005395 0.005426 0.005275 0.005124
std 0.000649 0.000632 0.000689 0.000638
min 0.004787 0.004831 0.004716 0.004537
25% 0.005142 0.005105 0.004985 0.004839
50% 0.005257 0.005283 0.005129 0.004985
75% 0.005438 0.005569 0.005364 0.005218
max 0.012633 0.012802 0.012514 0.012382
total 53.949636 54.256317 52.751914 51.244219
From the data (and the visuals), ./try-4.sh ran on average faster than the other 3 scripts. ./try-2.sh ran the slowest, which might highlight the inefficient regex matching from .*. ./try-3.sh ran slightly slower than ./try-4.sh, showing that it is better to run awk directly on the file as opposed to piping it in. Note however in order to make the data more readable outliers were not display. Data with outliers looks like this:
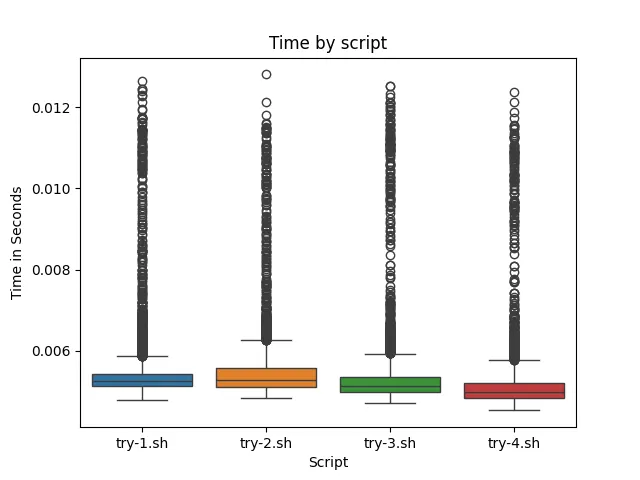
While the outliers help tell a story about the data, it seemed prudent to focus on the data within the IQR (interquartile range). In conclusion, if you want to save around .0002 to .0003 seconds, awk it up! This is slightly more performant than juggling a myriad of different bash commands.
I wanted to also test memory usage for the command, unfortunately I currently lack the skills to take a deep dive into memory analysis of a couple of bash scripts. (Perhaps a deep dive for another day)
If you are curious about any of the testing, I wrote a bash script to do it all. It can be found here: https://gist.github.com/AOrps/d0916ba18fc67c172476a1a34f7c2408
Note, that I am calling my Terraform / OpenTofu var-file .tfvar, the default files that OpenTofu looks for are terraform.tfvars and *.auto.tfvars. Which requires my command to look something like this:
tofu plan -var-file=.tfvar
Resources
- https://pilestone.com/pages/color-blindness-simulator-1 # trying to make the graphics used keep colorblindness in mind
- https://www.baeldung.com/ops/terraform-best-practices#1-using-variables-to-configure-a-provider
- https://owasp.org/www-community/attacks/Regular_expression_Denial_of_Service_-_ReDoS
- https://www.youtube.com/watch?v=DDe-S3uef2w
- https://blog.cloudflare.com/details-of-the-cloudflare-outage-on-july-2-2019/
- https://gist.github.com/AOrps/d0916ba18fc67c172476a1a34f7c2408